
Links
- Graduiertenkolleg 1026 "Conformational transitions in macromolecular interactions"
- Sonderforschungsbereich 610 "Protein-Zustände mit zellbiologischer und medizinischer Relevanz"
- ZIK HALOmem "Membrane protein structure and dynamics"
- ProNet-T3 "Protein-Kompetenznetzwerk-Halle: Tools, targets & therapeutics"
- Research group "Cryo Electron microscopy"
Contact
Prof. Dr. Milton T. Stubbs
Martin-Luther-Universität
Institut für Biochemie und Biotechnologie
phone: +49(0)345-5524901
fax: +49(0)345 5527014
stubbs@biochemtech.uni-hall...
Kurt-Mothes-Str. 3a
(Charles-Tanford-Proteinzentrum)
06120 Halle
Abteilung Physikalische Biotechnologie
Overview
Research interests within the X-ray group ‘Physikalische Biotechnologie’, established March 2002, lie in the structural biology of proteins of therapeutic relevance, with a particular focus on macromolecular conformational transitions. Projects fall roughly into three categories: natural product biosynthesis, signaling and membrane proteins, and the influence of protein flexibility on ligand affinity.
Natural product biosynthesis
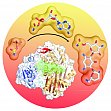
Parthier C, Görlich S, Jaenecke F, Breithaupt C, Bräuer U, Fandrich U, Clausnitzer D, Wehmeier UF, Böttcher C, Scheel D, Stubbs MT (2012) The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction. Angew Chem Int Ed Engl. 51, 4046-4052.
O-carbamoyltransferases, which modify a variety of natural products, exhibit an intriguing requirement for ATP. The structure of the 6”-O-carbamoyltransferase TobZ, responsible for biotransformation of the aminoglycoside antibiotic tobramycin to form nebramycin 5’, reveals two modular domains, each harboring an active site within a common ‘reaction chamber’. The C-terminal YrdC-like domain harbors a carbamoylphosphate binding site, occupation of which triggers Mg2+-ATP binding and subsequent formation of carbamoyladenylate seen here for the first time. The adenylated intermediate is channeled to a metal-ion-dependent nucleotide binding site within the N-terminal Kae1-like domain, where the carbamoyl moiety is transferred to the tobramycin acceptor hydroxyl group. Conservation of modules within components of the ubiquitous threonylcarbamoyladenosine-tRNA modification apparatus as well as the hydrogenase maturation factor HypF suggests analogous reaction sequences and points to an ancient origin of these proteins.
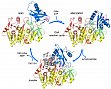
Yonus, H., Neumann, P., Zimmermann, S., May, J.J., Marahiel, M.A. and Stubbs, M.T. (2008) Crystal structure of DltA: Implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains. J. Biol. Chem. 283, 32484-32491.
DltA, the D-alanine:D-alanyl carrier protein ligase responsible for the initial step of lipoteichoic acid D-alanylation in gram positive bacteria, belongs to the adenylation domain superfamily, which includes also acetyl-CoA synthetase and the adenylation domains of non-ribosomal synthetases (NRPSs). The two step reaction catalyzed by these enzymes (substrate adenylation followed by transfer to the reactive thiol group of CoA or the phosphopantheinyl prosthetic group of peptidyl carrier proteins or PCPs) has been suggested to proceed via large scale rearrangements of structural domains within the enzyme. Structures of DltA reveal the determinants for DAla substrate specificity, and confirm that the PCP-activating domains are able to adopt multiple conformational states, in this case corresponding to the thiolation reaction. Comparisons of available structures have allowed us to propose a mechanism whereby small perturbations of finely balanced metastable structural states would be able to direct an ordered formation of NRPS products.
See also: Pfennig S and Stubbs MT (2012) Flexing and stretching in nonribosomal Peptide synthetases. Chem. Biol. 19, 167-169.
Signaling and membrane proteins
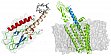
Parthier, C., Kleinschmidt, M., Neumann, P., Rudolph, R., Manhart, S., Schlenzig, D., Fanghänel, J., Rahfeld, J.-U., Demuth, H.-U. and Stubbs, M.T. (2007) Crystal structure of the incretin-bound extracellular domain of a G-protein coupled receptor. PNAS 104, 13942-13947.
Incretins, endogenous polypeptide hormones released in response to food intake, potentiate insulin secretion from pancreatic b-cells after oral glucose ingestion (incretin effect). This response is signaled by the two peptide hormones GIP (glucose-dependent insulinotropic polypeptide or gastric inhibitory polypeptide) and GLP-1 (glucagon-like peptide 1) through binding and activation of their cognate class 2 G-protein coupled receptors (GPCRs). As the incretin effect is lost or significantly reduced in patients with type 2 diabetes mellitus, GLP-1 and GIP have attracted considerable attention for their potential in anti-diabetic therapy. A paucity of structural information precludes a detailed understanding of the processes of hormone binding and receptor activation, hampering efforts to develop novel pharmaceuticals. The crystal structure of the complex of human GIP receptor extracellular domain (GIPR-ECD) with its agonist, the incretin GIP1-42 shows that the hormone binds in an a-helical conformation in a surface groove of the ECD largely via hydrophobic interactions. The N-terminal ligand residues would remain free to interact with other parts of the receptor. Thermodynamic data suggest that binding is concomitant with structural organization of the hormone, resulting in a complex mode of receptor-ligand recognition. The presentation of a well-structured, a-helical ligand by the ECD is expected to be conserved among other hormone receptors of this class.
See also: Parthier, C., Runge, S., Rudolph, R. and Stubbs M.T. (2009) Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem. Sci. 34, 303-310.

Neumann, P., Lieber, D., Meyer, S., Dautel, P., Kerth, A., Kraus, I., Garten, W. and Stubbs, M.T. (2009) Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals single-stranded RNA binding properties. PNAS 106, 3710-3715.
Borna disease virus (BDV) is a neurotropic enveloped RNA virus that causes a non-cytolytic, persistent infection of the central nervous system in mammals. BDV belongs to the order Mononegavirales, which also includes the negative strand viruses Ebola, Marburg, vesicular stomatitis, rabies and mumps and measles. BDV-M, the matrix protein of Borna disease virus, is the smallest matrix protein (16.2 kDa) among the negative strand viruses. Matrix proteins play a critical role in virus assembly and budding, mediating the interaction between the viral capsid, envelope and glycoprotein spikes, and are as such responsible for the structural stability and the individual form of virus particles. The three-dimensional structure of BDV-M, the first full length matrix protein structure from a negative-stranded non-segmented RNA virus, indicates that the BDV-M monomer exhibits structural similarity to the N-terminal domain of the Ebola matrix protein VP40, while the surface charge of the tetramer provides clues to the membrane association of BDV-M. Additional electron density in the crystal reveals the presence of bound nucleic acid, interpreted as cytidine-5’-monophosphate. The heterologously expressed BDV-M copurifies with and protects single-stranded RNA oligonucleotides of a median length of 16 nucleotides. The results presented here show that BDV-M would be able to bind RNA and lipid membranes simultaneously, expanding the repertoire of matrix protein functionalities.

Hoffmann, A., Funkner, A., Neumann, P., Juhnke, S., Walther, M., Weininger, U., Schierhorn, A., Balbach, J., Reuter, G. and Stubbs, M.T. (2008) Biophysical characterisation of refolded Drosophila Spätzle, a cystine knot protein, reveals distinct properties of three isoforms. J. Biol. Chem. 283, 32598-32609.
The Drosophila Spätzle protein, involved in the embryonic development of the dorsal-ventral axis and in the adult immune response, is expressed as a pro-protein and is activated by the serine proteinases Easter or Spätzle Processing Enzyme. Proteolytic cleavage generates a 106 amino acid C-terminal fragment C106 homologous to the mature form of nerve growth factor NGF, a cystine knot protein. Through alternative splicing, the Spätzle gene encodes for several isoforms that with one exception (the ‘pro-peptide isoform’) share C106 but differ in the pro-sequence. Three isoforms have been expressed recombinantly in E. coli strains. The pro-peptide isoform could be expressed in soluble form and is unstructured according to CD and NMR measurements. Dimeric full length Spätzle isoforms have been refolded from insoluble inclusion bodies and are able to rescue Spätzle-deficient embryos. While the two full length isoforms exhibit similar far-UV CD spectra, large differences in tryptophan fluorescence quenching by the respective pro-parts are observed. Both full length isoforms exhibited highly cooperative folding transitions. Proteolytic digestion using trypsin resulted in C106, whose unfolding exhibits lower thermodynamic stability and cooperativity compared to the full length proteins. The structure of C106 reveals a T-shaped dimer with significant differences to NGF and a deep internal cavity. Substantial b-sheet formation is observed between the two monomers, while a long loop containing the single tryptophan residue is disordered in the crystals. Our results suggest that the pro-peptides stabilize the tertiary structure of the ‘mature’ Spätzle cystine knot.
Protein - ligand interactions
In the field of "rational drug design", we try to fuse the experimental structural data and thermodynamic measurements of protein ligand interactions in a computational framework to advance the drug discovery process.
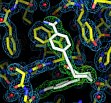
Rauh, D., Klebe, G. and Stubbs, M.T. (2004) Understanding protein-ligand interactions: The price of protein flexibility. J. Mol. Biol. 335, 1325-1341.
Carrillo DR, Parthier C, Jänckel N, Grandke J, Stelter M, Schilling S, Boehme M, Neumann P, Wolf R, Demuth HU, Stubbs MT* and Rahfeld JU (2010) Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus. Biol Chem. 391, 1419-1428.
Ruiz-Carrillo D, Koch B, Parthier C, Wermann M, Dambe T, Buchholz M, Ludwig HH, Heiser U, Rahfeld JU, Stubbs MT*, Schilling S and Demuth HU (2011) Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center. Biochemistry 50, 6280-6288.
Schöpfel M, Tziridis A, Arnold U and Stubbs MT (2011) Towards a restriction proteinase: construction of a self-activating enzyme. Chembiochem 12, 1523-1527.
Cryo electron microscopy

Han BG, Walton RW, Song A, Hwu P, Stubbs MT, Yannone SM, Arbelaez P, Dong M, Glaeser RM. Electron microscopy of biotinylated protein complexes bound to streptavidin monolayer crystals. J Struct Biol. 2012 May 11.
Chemical biotinylation of protein complexes followed by binding to two-dimensional (monolayer) crystals of streptavidin is shown to be an effective way to prepare cryo EM specimens from samples at low protein concentration. Three different multiprotein complexes are used to demonstrate the generality of this method. In addition, native thermosomes, purified from Sulfolobus solfataricus P2, are used to demonstrate that a uniform distribution of Euler angles is produced, even though this particle is known to adopt a preferred orientation when other methods of cryo EM specimen preparation are used.



